Active data enrichment by learning what to annotate in digital pathology
March 11, 2024
Abstract: Our work aims to link pathology with radiology with the goal to improve the early detection of lung cancer. Rather than utilising a set of predefined radiomics features, we propose to learn a new set of features from histology. Generating a comprehensive lung histology report is the first vital step towards this goal. Deep learning has revolutionised the computational assessment of digital pathology images. Today, we have mature algorithms for assessing morphological features at the cellular and tissue levels. In addition, there are promising efforts that link morphological features with biologically relevant information. While promising, these efforts mostly focus on narrow well-defined questions. Developing a comprehensive report that is required in our setting requires an annotation strategy that captures all clinically relevant patterns specified in the WHO guidelines. Here, we propose and compare approaches aimed to balance the dataset and mitigate the biases in learning by automatically prioritising regions with clinical patterns underrepresented in the dataset. Our study demonstrates the opportunities active data enrichment can provide and results in a new lung-cancer dataset
annotated to a degree that is not readily available in public domain.
Read more here
Important: DART Programme Data Email Address Update
March 17, 2025
Thank you to all the sites which are sending data to the DART Programme. Please note that our email address…
Update on the NHS Lung Cancer Screening Programme
March 4, 2025

DART at OxCODE
January 29, 2025
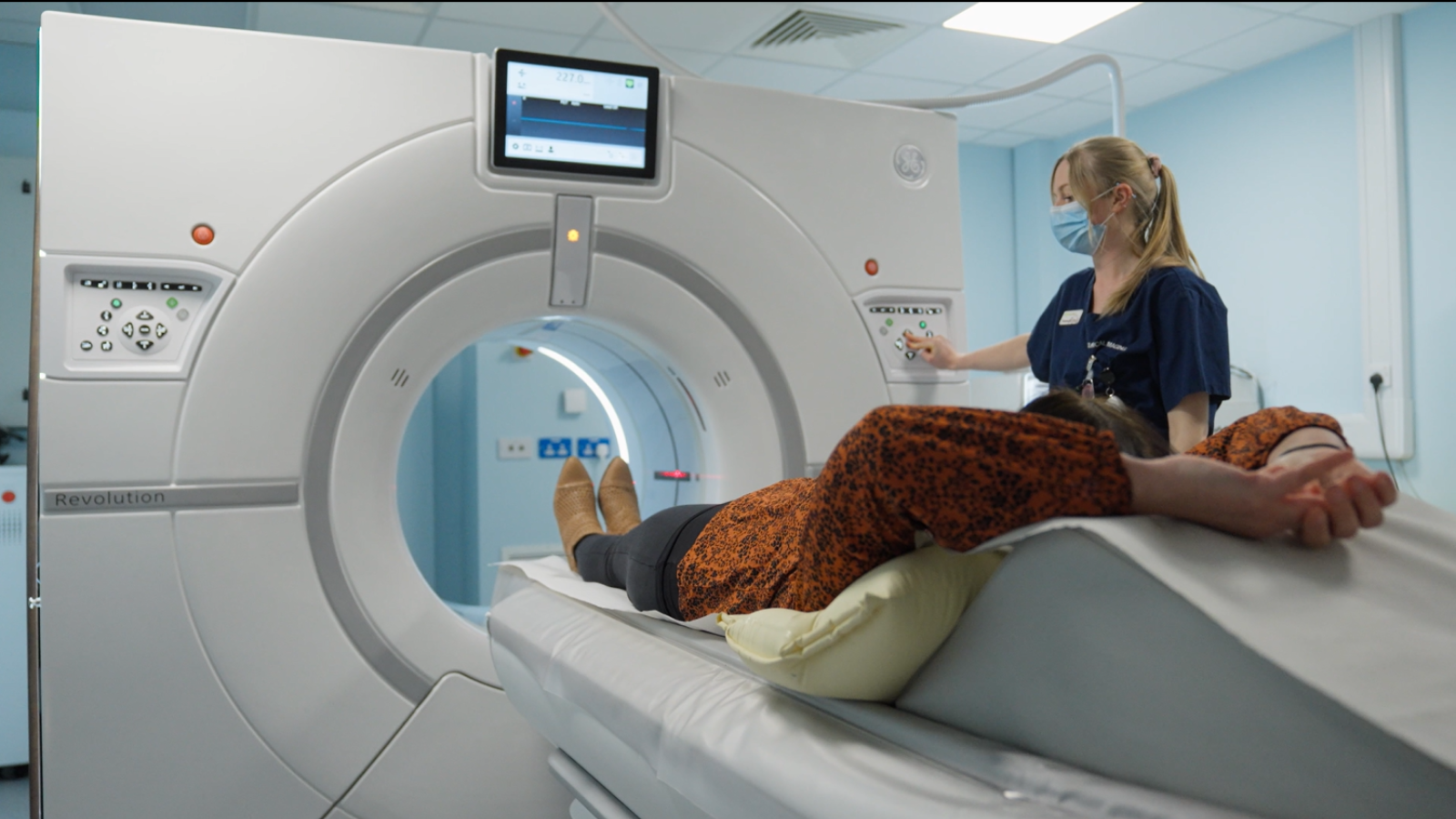
Unlocking the future of lung cancer screening: How DART is redefining early diagnosis
October 23, 2023